Imaginary Orangutans and the Baldwin Effect
Posted: 11/07/2024Most people are familiar with the notion of natural selection, a process by which genetic change takes place within a population over many generations based on fitness. The driving principle of natural selection is that good genes will outlive and outproduce bad genes eventually resulting in a well adapted population. In this blog I'll attempt to address the effect individual learning has on evolution, a phenomenon known as the Baldwin Effect, and elaborate on a simulation model for observing this effect.
The existence of the Baldwin Effect has long since been fairly well proven and was first brought to my attention by Geoffrey Hinton and Steven Nowlan's paper, How Learning Can Guide Evolution. Even at the time of its writing (1987) the existence of the Baldwin Effect was not particularly up for debate having first been proposed 100 years earlier in 1886 (by the eponemous James Baldwin) and eventually accepted as part of the Modern Synthesis. However, their contribution of a simple model which is still capabale of demonstrating the Baldwin Effect provides a good platform for going slightly beyond intuition and persuading yourself of its existence.
Hinton and Nowlan's model is not without flaw, it is extremely abstract. To try and alleviate this I'm going to try and break this post into two parts. First, a simplified example designed to build basic intuition about the Baldwin Effect. Second, an outline of Hinton and Nowlan's model which builds upon this intuition.
Part 1: An Example
Lets imagine a population of Orangutans that can vary genetically based on two genes:
- Gene 1, Spotting: Ability to spot durian trees
- Gene 2, Digesting: Ability to effectively digest durian
And then assume that each gene has alleles (forms that gene may take, e.g. eye colours) ranging from "weak" to "strong". It's fairly obvious that some combinations of these two genes will be the best, lets simplify them in a table:
| Strong Spotter | Weak Spotter | |
| Strong Digestor | A.) Very healthy! Lots of durian, lots of digestion. | B.) Okay. Not a lot of durian, great digestion. |
| Weak Digestor | C.) Okay. Lots of durian, bad digestion. | D.) Oh no. Not a lot of durian, bad digestion. |
So an Orangutan who is awful at finding durian trees but amazing at digesting durians would be type B, lets call this (rather unfortunate) orangutan Beni. This seems a pretty unlucky combination of genes and certainly if genes alone where responsible for behaviour Beni would have a pretty bad time exploiting his excellent durian-digesting abilities. But we know this isn't true, throughout his life Beni can learn to become a better durian tree spotter. Even if he only reaches an adequate level of durian-spotting he's able to get so much nutrients from each durian that he ends up having a reproductively successful life. This is great news. Beni's durian digestion gene, which might otherwise have been lost, is preserved in the population!
This is, roughly, the notion of the Baldwin Effect. Beni's ability to learn behaviour had a direct effect on the genetic makeup of the population. It is worth noting that orangutans display not only individual but social learning with knowledge being passed down from parents to offspring. This type of cultural or memetic evolution will be ignored for now to better focus the discussion on demonstrating the Baldwin Effect in code. Maybe it will appear in a later blog!
If you've read up to this point then I hope, at the very least, the promise of this blogs title is somewhat fulfilled. So if you are satisfied just to have learned what the Baldwin Effect is and perhaps gained some new-found perspective on the complexities of evolutionary processes then feel free to skip to the conclusions and stop here!
Part 2: Hinton and Nowlan's Model
Now we've got an idea of what the Baldwin Effect is we can address the question of how best to design an evolutionary model which can demonstrate it. Hinton and Nowlan present a model which is just complex enough to do just this whilst still being easy to implement and explain:
- Instead of just two genes, each member of the population has 20 genes
- Each gene has three alternate forms: 0, 1 and ? and corresponds to an edge in a small Neural Network1
- 0s represent deactivated edges along which neural signals are blocked
- 1s represent activated edges along which neural signals can pass
- ?s represent edges which may be either active or inactive
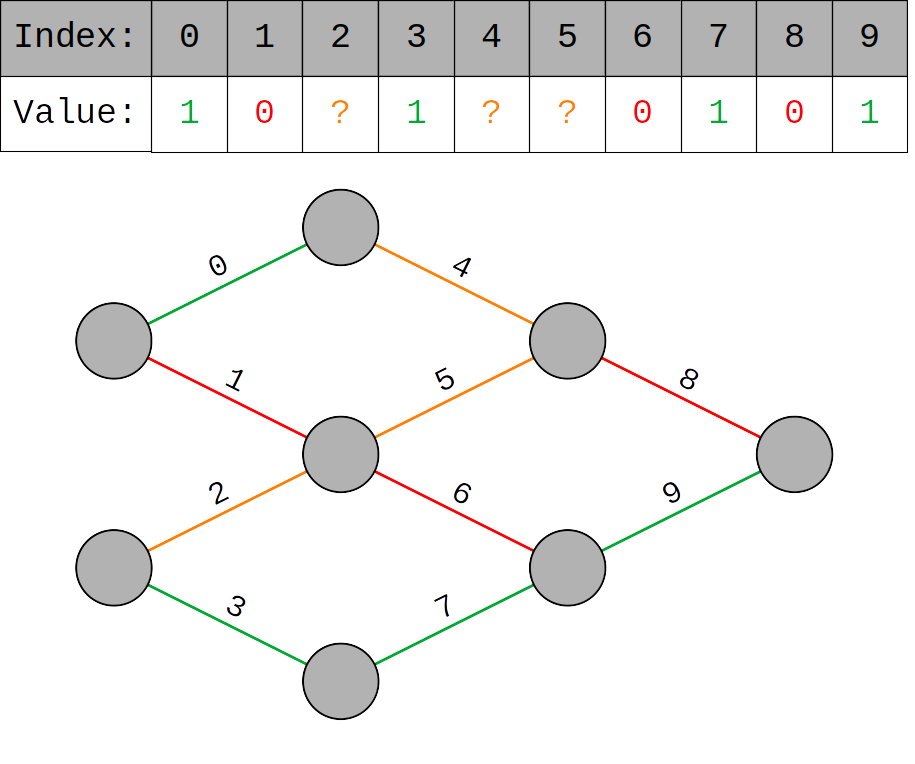
The 0 and 1 genes cannot be altered but ? genes can essentially be chosen by the organism. This assignment of ? genes is the analog for individual learning and thus the learning task of each organism is to find the assignment of ? genes which maximizes their performance.
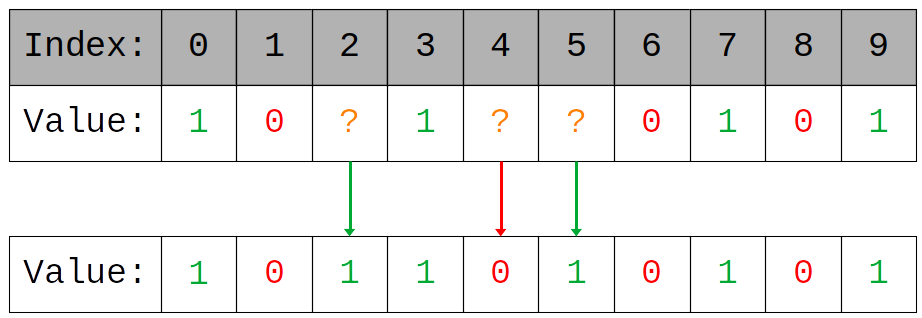
For their experiment Hinton and Nowlan suppose that the network performs best when fully activated, a genetic sequence of only 1 genes. This extreme choice of optima is intentional and was chosen to make demonstration easier. Now we have a genetic model which facilitates some notion of individual learning we need an environment to embed this model in, an Evolutionary Game.
The Evolutionary Game Framework
Evolutionary Games extend the standard Game Theoretic model of competing organisms (henceforth "players") within a population to allow for multiple generations of players whose strategies may evolve over time. Each player participates in a series of rounds receiving some reward based on how successful they are in each round. Each sequence of rounds forms a generation. After each generation a reproductive process is applied determining the genetic makeup of the next generation. Lets elaborate on each of these aspects (rounds, generations and reproductive processes) so we can better understand the model as a whole.
Rounds
In each round a player applies their strategy and receives some reward based on how good that strategy is. So for our model a strategy is a player's genetic sequence and a player wont receive any reward unless their strategy is exactly the optima, all 1s. For simplicity we'll give 1 reward in this case and 0 otherwise. Our players will employ a very simple (and very dumb) random guessing strategy where they assign their ?s at random until they get some reward, after this point they never change their strategy again.
Generations
A generation is a series of rounds, between each round the players can alter their strategies (assign ?s). Reward accumulates over each generation so the earlier a player learns the correct sequence the better, a player who only figures out the best sequence on round 99 out of 100 will get just 1 reward for the whole generation.
Reproductive Process
After each generation the reward received by a player is used to determine how likely they are to have offspring in the next generation. To give an example, if the total reward earned by all players in a generation is 100 and a single player earns 50 reward, we would expect some of their genes to appear in 50% of the next generations players. Players reproduce in pairs via splicing of their genetic sequences where a random cutoff determines how much of each parents' sequence is present in the child. It's worth remembering that parent selection and reproduction is still a random process, so the best performing player is not guaranteed to reproduce, nor is the worst performing guarenteed not to.
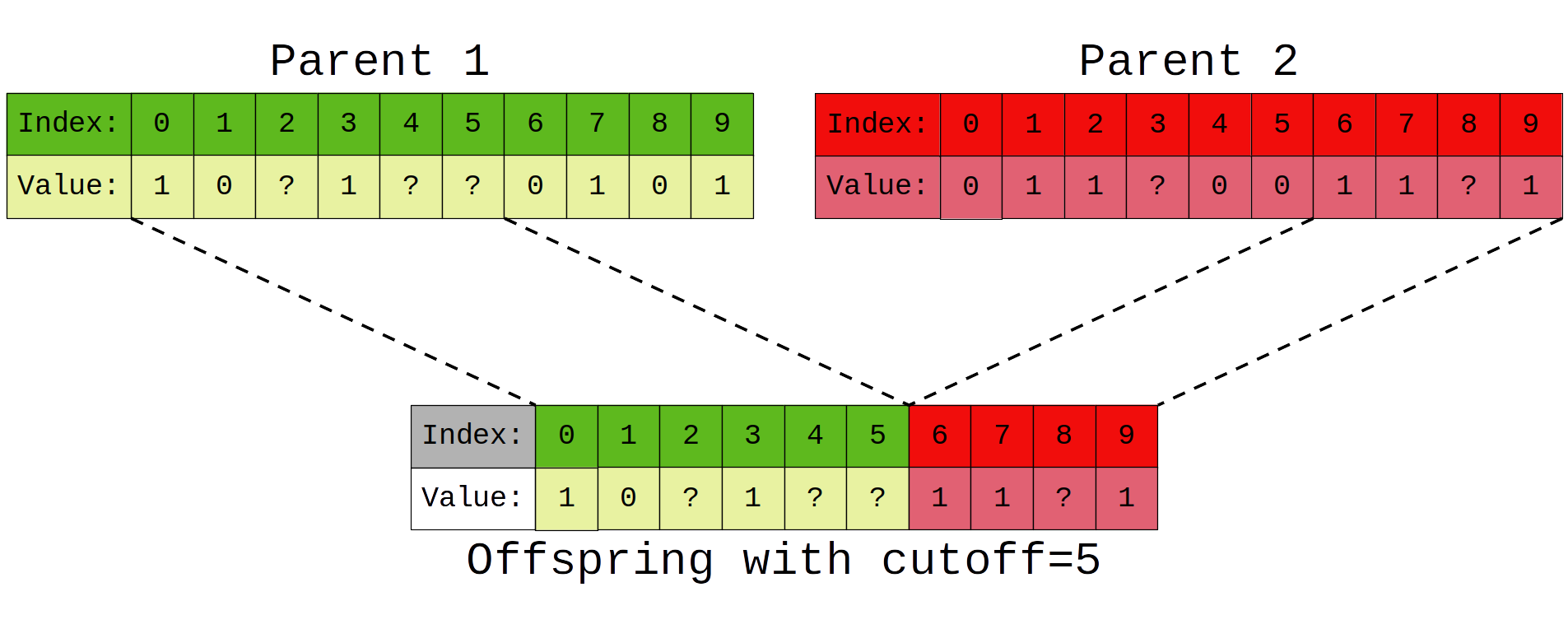
So to summarise, our evolutionary game goes something like this:
- Generate an initial population with random genetic sequences
- Players submit ? assignments each round until the generation ends
- The reproductive process is applied based on the rewards for this generation
- Go back to step 2 with the newly produced generation
Using the Model, Experiments
So what are the results of actually simulating this model? Well, both of the following graphs show a population of 1000 players simulated over 30 generations of 1000 rounds with a genetic sequence length of 20. These parameters are taken from one of Hinton and Nowlans own experiments (found here). The first graph shows the results when starting from an initial gene pool, made up of 50% ? genes and 25% each of 0 and 1 genes. The second graph shows a population who have no learnable (?) genes:
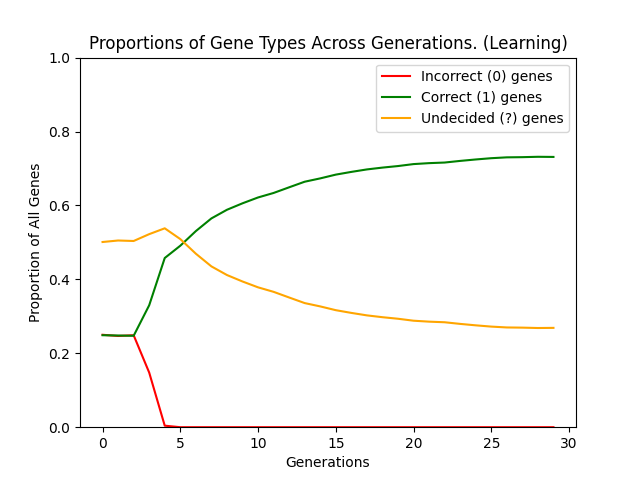
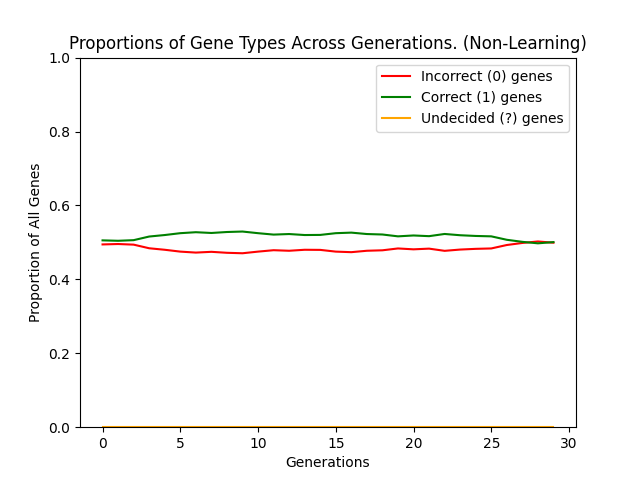
The difference is quite stark (as you might expect) and in fact the non-learning population never discovers the optimal sequence. On the other hand the learning population is able to find the optima with relative ease and then stabilise within a genetic landscape where the optima can be consistently achieved. It is essentially this ability to reach the optima not simply by finding the single genetic optima but by reaching a genetic region of "near-optimal" sequences which can then be exploited which smooths the genetic search space, causing the Baldwin Effect.
Conclusions and Further Resources
I hope at the very least that this post helped to introduce you to some new evolutionary ideas. Darwin's theory is an extremely important scientific contribution and even a very basic understanding of it offers a pretty solid perspective on evolution. That being said, as with most things, evolution is quite complex and I think exploring different facets and forms of evolution helps to develop a better appreciation for just how impressive and fascinating it can be.
They say you don't really understand something until you've implemented it and to that end I made an effort to produce a minimal implementation of this model in Python, you can find it here. Additionally, many of the resources used in this post are taken from this presentation which I produced on the same topic.
1. An understanding of NNs or Deep Learning is not required to understand this post. An NN was chosen by Hinton and Nowlan likely because of its generalisability and (at the time more relevant) supposed relatedness to biological brain structure. It could been almost any structure or policy encodable by a string of only 3 symbols.↩